Brief Summary
On this page, we mainly describe considerations made when designing the system. We also describe how this system can be utilized in the future by society and issues that arise with human beings and society.
DNA Design Innovation
In this section, we describe some points of devises for realizing the DNA circuit described above. These depart from the subject of this project, however they have important meanings for realization.
1. Application of Mismatched Base Pair
We described the features of mismatched base pairs and an application example.
In designing DNA, it is necessary to accurately predict the DNA’s behavior and design base sequences.
However, as the DNA system becomes more complex, greater deviation from the expected results will be observed.
To solve this problem, special secondary structures, such as a hairpin loop or bulge loop (Figure 1), are typically used.

However, bot the benefits and defects caused by using the method must be considered. For example, for defects in the bulge loop, as the base sequence of a part of the bulge increases in length, the secondary structure becomes more unstable (Figure 2).
Also, there is a possibility that a leakage reaction may occur from the single strand portion of the bulgeil.
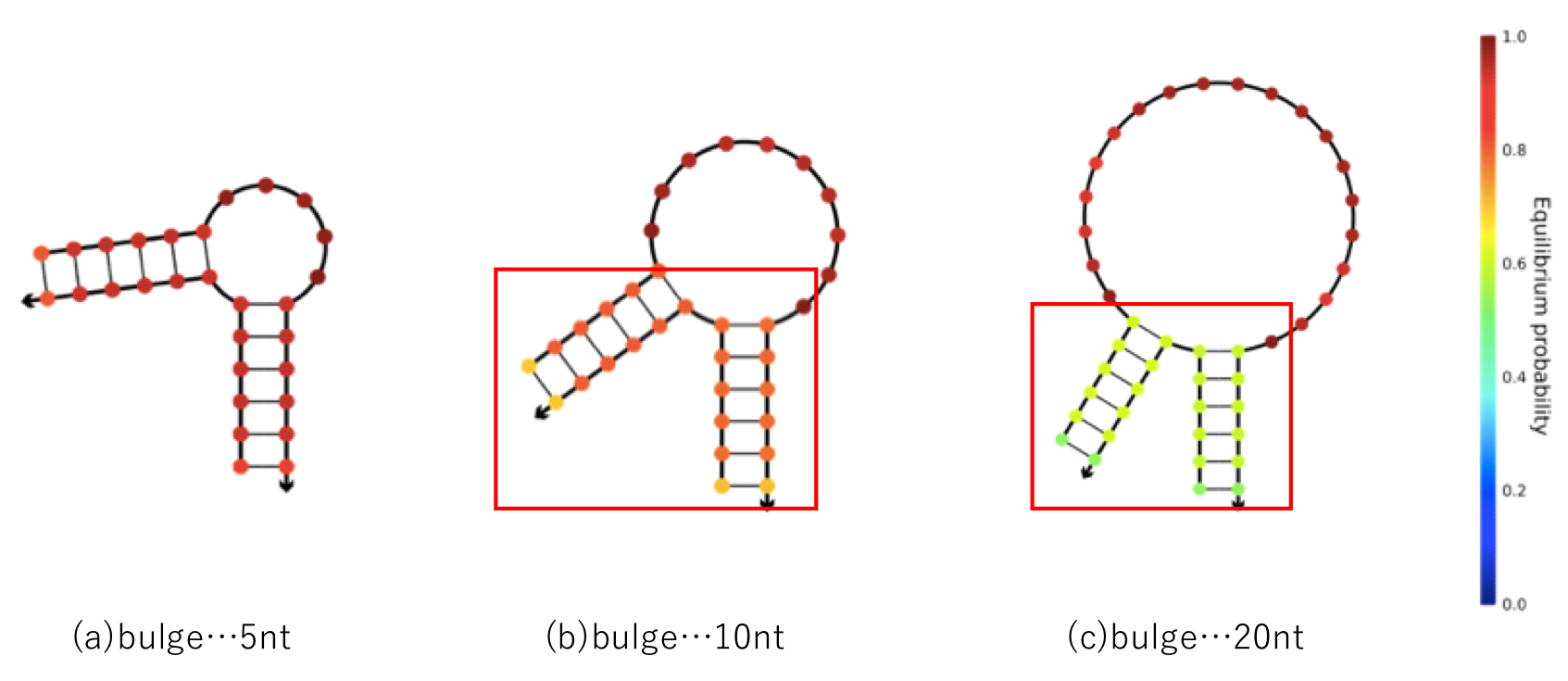
Therefore, these features are used for different purposes according to the goal. We use the new method ‘Mismatched base pair’ to control the DNA as accurately as possible. Mismatched GT base pairs can solve a variety of problems. The primary advantage of mismatched base pairs is the simplicity of the mechanism. In other methods, detailed knowledge and skills are required to minimize unintentional behaviors of DNA. Additionally, the circulation must be redesigned many times as it becomes more complex. However, for mismatched base pairs, only parts that may not behave as expected are identified mismatched base pairs are added. However, there are some limitations to this method, such as the long time required to introduce these base pairs into the experiments. Accordingly, various patterns of mismatched base pairs must be tested. Additionally, there are no simulation tools using mismatched base pairs. Because mismatched base pairs can be used in various applications, the development of mismatched base pair technology will greatly contribute to the development of molecular robotics.
2. Strength of DNA Binding
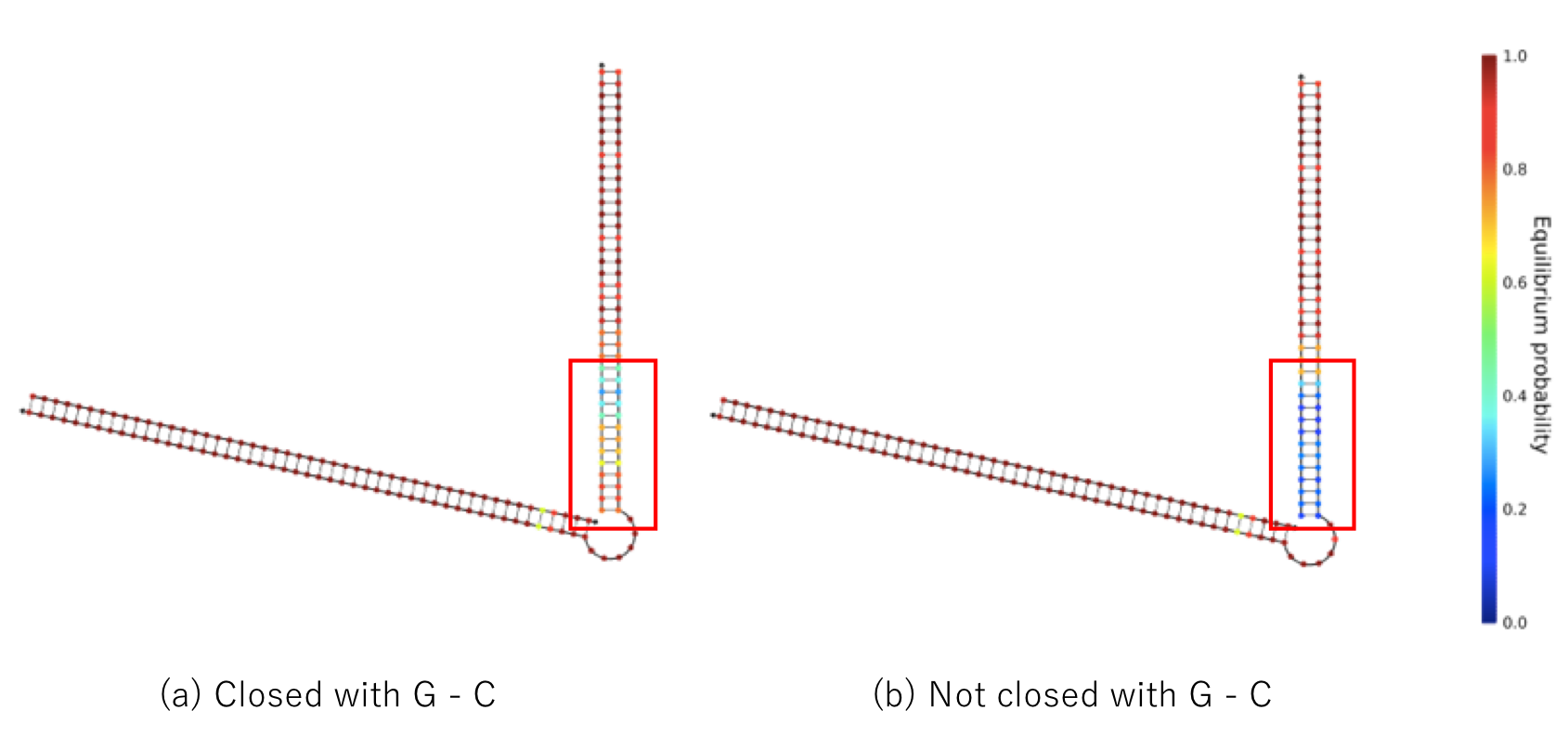
We mentioned the strength of DNA binding. In this project, we used the T7 promoter sequence, but intentionally added G-C bonds at both ends of the chain because G-C bonds are stronger than A-T bonds, and double-stranded DNA is stable among Watson Crick base pairs. Particularly, this strategy is necessary because the binding force of double-stranded DNA is weakened by adding mismatched base pair. Figure 3 compares the case in which a G-C bond is added to the edge and a case in which it is not added. We found that double-stranded DNA was more stable when a G-C bond was present. This prevents unintentional behavior caused by instability of the secondary structure.
3. Delay of the Seesaw Gate
We used a seesaw gate to delay the output time. The most common method of slowing the reaction rate of DNA is to change the length of the toehold. Recently, regulation of the reaction rate by mismatched base pair has been reported. The major advantage of adjusting the time using the seesaw gate is that adjustment can be conducted by simply changing the density of the threshold gate. For a toehold or mismatched base pair, it is necessary to determine the reaction rate at the time of base sequence design; therefore, to adjust the time according to the application, it is efficient to use the seesaw gate.
Future
This section describes the applications of the ATP controller.
Particularly, we focused on cancer treatment. Currently, the main cancer treatments are radiation therapy and anticancer drugs, but there are some limitations to these therapies.
These treatments damage not only cancer cells, but also normal cells.
To ensure that only cancer cells are damaged, we focused on the differences in energy generation between normal cells and cancer cells. First, we explain how normal cells generate energy. Carbohydrates such as bread, rice, and noodles are decomposed into glucose. Cells incorporate glucose, which is used to generate ATP in 3 steps ((1) glycolysis, (2) TCA cycle, and (3) electron transport chain).
The first step is glycolysis. In the cytoplasm, glucose is decomposed into pyruvate. A small amount of ATP is generated by glycolysis. In the presence of oxygen, pyruvate is incorporated into the mitochondria. The second step is the TCA cycle. In the mitochondria, pyruvate is converted to acetyl-CoA, which is decomposed into NADH and FADH2 in the TCA cycle. As in glycolysis, a small amount of ATP is generated. The third step is the electron transfer chain. In this system, a large amount of ATP is generated. Therefore, normal cells can generate ATP efficiently. Cancer cells do not use mitochondria based on research by Warburg in 1926. Thus, ATP is generated only through glycolysis in cancer cells, which is an inefficient process[13][14].
.png)
Figure 4: Metabolism of Normal Cells
.png)
Figure 5: Metabolism of Cancer Cells
Cancer cells require more ATP than normal cells because they undergo rapid division.
However, glycolytic is not efficient for generating ATP.
Some cancer treatments involve inhibition of glucose or glycolysis. Thus, absorbing ATP greatly influences cancer cells.
.png)
Figure 6: Metabolism of Normal Cells after Administration
.png)
Figure 7: Metabolism of Cancer Cells after Administration
Finally, we propose that the ATP controller can be used to assist in cancer treatment because of its effects on metabolism.
ELSI
We are developing a system for controlling the amount of ATP in the cell signaling system using DNA. We are conducting verification experiments in vitro, and the effect on the human body has not been confirmed. However, if medicines using this system are generated in the future and administered in vivo, adverse effects on the human body can be prevented by reducing ATP levels.
Because criminal investigation and military applications using these adverse effects are predicted, it is necessary to evaluate the use of this method in vivo. Although this system uses DNA, it does not contradict regulations of the Cartagena Protocol because the project was domestically established and is currently in the research and development stage; additionally, genetic modification is not required. However, it is expected that DNA circuits will be administered to genetically modified organisms in the future. To administer DNA, it is necessary to consider the influence on the genes of organisms. Thus, efforts much be made to comply with the Cartagena Protocol and other laws.
References
[1]Boris N. Kholodenko, Oleg V. Demin, Gisela Moehren, and Jan B. Hoek (1999) "Quantification of Short Term Signaling by the Epidermal Growth Factor Receptor" J Biol Chem, 274(42), 30169-30181
[2]D. Zhan and E. Winfree (2009) "Control of DNA strand displacement kinetics using toehold exchange" J. Am. Chem. Soc., 131(47), 17303-17314
[3]S. Kobayashi, K. Yanagibashi, K. Fujimoto, K. Komiya and M. Hagiya (2014) "Analog DNA computing devices toward the control of molecular robots", Workshop on Self-organization in Swarm of Robots: from Molecular Robots to Mobile Agents (WSSR 2014)
[4]Lulu Qian and Erik Winfree (2011) "A simple DNA gate motif for synthesizing large-scale circuits" J R Soc Interface, 8(62), 1281–1297, doi:10.1098/rsif.2010.0729
[5]Lulu Qian and Erik Winfree (2011) "Neural Network Computation with DNA Strand Displacement Cascades" Nature, 475, 368–372
[6]Yuki Yoshida, Takashi Nakakuki (2017) "Design tool of DNA base sequences for molecular circuits" Proc. of 17th International Conference on Control, doi: 10.23919/ICCAS.2017.8204313
[7]NUPACK: http://www.nupack.org
[8]Matlab: https://jp.mathworks.com/products/matlab.html
[9]Nakakuki T, Yumoto N, Naka T, Shirouzu M, Yokoyama S, Hatakeyama M. (2008) "Topological analysis of MAPK cascade for kinetic ErbB signaling." PLoS One, 3(3):e1782. doi: 10.1371/journal.pone.0001782
[10]Tong Li, Hoi Hung Ho, Maribeth Maslak, Charlie Schick, and Craig T. Martin (1996) "Major Groove Recognition Elements in the Middle of the T7 RNA Polymerase Promoter" Biochemistry, 35(12), 3723
[11]MINQING RONG, BIAO HE, WILLIAM T. MCALLISTER, AND RUSSELL K. DURBIN (1998) "Promoter specificity determinants of T7 RNA polymerase" Proc. Natl. Acad. Sci. USA, 95(2), 516
[12]Reinhart Heinrich, Benjamin G. Neel, and Tom A. Rapoport (2002) "Mathematical Models of Protein Kinase Signal Transduction" Molecular Cell, 9(2), 957-970
[13]Ran Wei, Limin Mao, Ping Xu, Xinghai Zheng, Robert M. Hackman, Gerardo G. Mackenzie and Yuefei Wang (2018) "Suppressing glucose metabolism with epigallocatechin-3-gallate (EGCG) reduces breast cancer cell growth in preclinical models" Food Funct, doi: 10.1039/C8FO01397G
[14]Gang Wang, Yifeng Li, Zeyu Yang, Weina Xu, Yifan Yang, Xiaodong Tan (2018) "ROS mediated EGFR/MEK/ERK/HIF-1α Loop Regulates Glucose metabolism in pancreatic cancer" Biochemical and Biophysical Research Communications, 500(4), 873-878